What’s a netCDF file?
NetCDF (Network Common Data Form) is a set of software libraries and self-describing, machine-independent data formats that support the creation, access, and sharing of array-oriented scientific data. The project homepage1 is hosted by the Unidata program at the University Corporation for Atmospheric Research (UCAR). They are also the chief source of netCDF software, standards development, updates, etc. The format is an open standard. NetCDF Classic and 64-bit Offset Format are an international standard of the Open Geospatial Consortium.2
The way I see a netCDF file is that it’s a matrix that usually contains environmental data across time and location, plus extra information (called metadata) such as where does the dataset comes from, or its resolution.
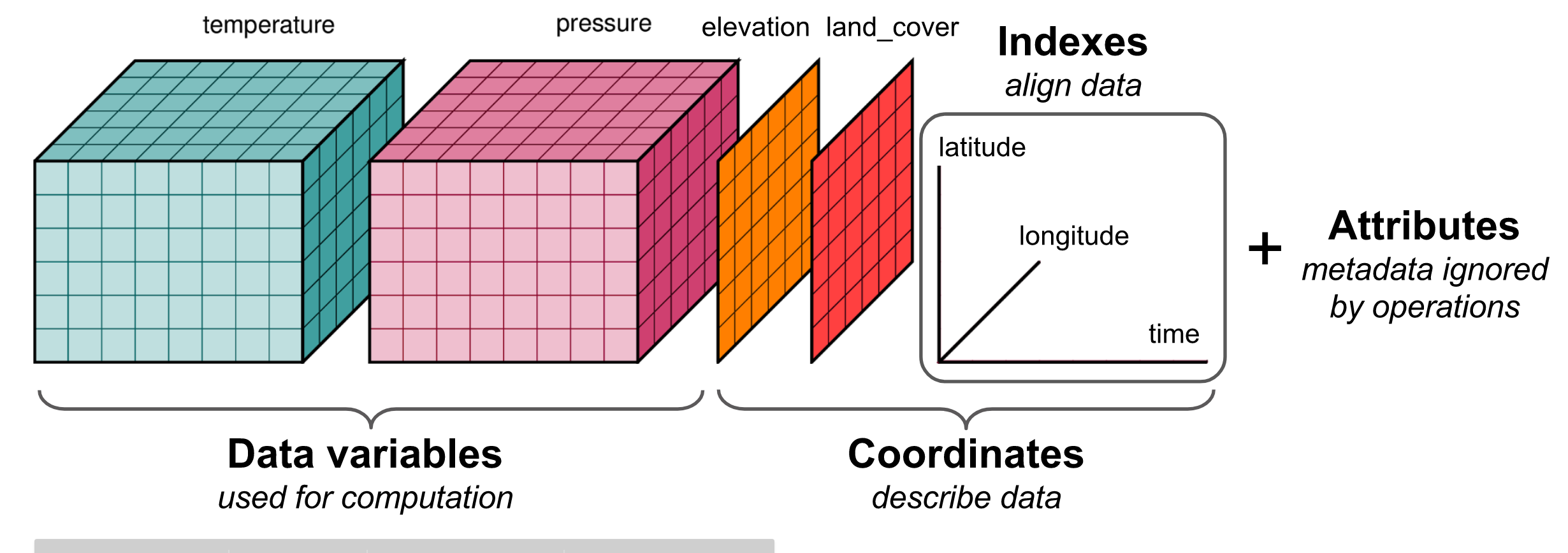
How to open it?
Super simple, using the tidync
package! Let’s say you have a netcdf file called
global-reanalysis-phy-001-031-grepv2-daily_1639765258612.nc,
here is what you have to do to import this file:
# first load `tidync` package
library(tidync)
# then import your file
df_nc <- tidync("../inst/extdata/copernicus/global-reanalysis-phy-001-031-grepv2-daily_1639765258612.nc")
# flatten the multidimensional array
df <- df_nc %>%
# extract NetCDF data as an expanded table
hyper_tibble() %>%
# not mandatory, but here is the code to convert to a data.table
setDT()
# print the first 10 rows
df[sample(nrow(df), 10), ] %>%
sable(
caption = "First 10 rows of the flatten netCDF file",
digits = 2
)| uo_cglo | uo_glor | vo_oras | uo_foam | vo_glor | uo_oras | vo_cglo | vo_foam | longitude | latitude | depth | time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.05 | 0.20 | -0.12 | 0.11 | -0.06 | 0.10 | -0.02 | -0.13 | -140.00 | 51.00 | 0.51 | 25148 |
| 0.06 | 0.04 | 0.04 | 0.10 | 0.03 | 0.01 | -0.04 | -0.21 | -127.00 | 43.50 | 0.51 | 25161 |
| -0.07 | -0.17 | -0.06 | -0.23 | -0.11 | -0.17 | -0.16 | -0.08 | -130.25 | 36.75 | 0.51 | 25400 |
| 0.03 | -0.06 | -0.03 | -0.01 | -0.12 | 0.01 | 0.05 | -0.11 | -131.75 | 27.75 | 0.51 | 25078 |
| -0.03 | -0.14 | -0.08 | 0.00 | 0.01 | -0.04 | -0.09 | -0.04 | -156.25 | 30.00 | 0.51 | 25363 |
| 0.04 | 0.02 | 0.03 | 0.06 | 0.04 | 0.04 | -0.02 | -0.07 | -136.00 | 35.50 | 0.51 | 25382 |
| 0.01 | -0.01 | 0.11 | 0.00 | -0.07 | 0.05 | -0.02 | -0.03 | -152.75 | 48.25 | 0.51 | 25284 |
| 0.15 | 0.17 | 0.04 | 0.07 | 0.07 | 0.07 | 0.13 | 0.02 | -149.25 | 28.25 | 0.51 | 25240 |
| -0.02 | -0.04 | 0.00 | -0.08 | -0.02 | -0.04 | -0.02 | -0.05 | -127.00 | 43.50 | 0.51 | 25314 |
| 0.11 | 0.16 | -0.11 | 0.10 | 0.00 | 0.11 | -0.08 | -0.06 | -160.50 | 40.75 | 0.51 | 25164 |
Now we can see that the time is not in a proper format. The first
thing to do is to look into the metadata to see if we could find some
kind of explanation. To do that, we’re going to use the
ncmeta package, developped for this purpose.
# load ncmeta library
library(ncmeta)
# then we use the function nc_atts to access attributes of times
print(
nc_atts(
"../inst/extdata/copernicus/global-reanalysis-phy-001-031-grepv2-daily_1639765258612.nc",
"time"
) %>%
# we want information on the unit for time column
dplyr::filter(name == "units") %>%
pull(value)
)## $units
## [1] "days since 1950-01-01 00:00:00"The column time seems to be the number of days since
1950-01-01. Let’s use that information to format the
time column in a proper way.
# convert time into a readable format
df[, time := as.Date(time, origin = as.Date("1950-01-01"))]
# print
df[sample(nrow(df), 10), ] %>%
sable(
caption = "First 10 rows of the flatten netCDF file, with reformat time column",
digits = 2
)| uo_cglo | uo_glor | vo_oras | uo_foam | vo_glor | uo_oras | vo_cglo | vo_foam | longitude | latitude | depth | time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| -0.01 | 0.04 | 0.01 | 0.04 | -0.08 | -0.04 | -0.01 | 0.02 | -141.50 | 50.25 | 0.51 | 2019-01-02 |
| 0.02 | 0.11 | 0.03 | 0.10 | 0.04 | 0.00 | 0.08 | 0.06 | -138.50 | 27.50 | 0.51 | 2018-07-25 |
| 0.03 | 0.01 | -0.01 | 0.09 | 0.00 | 0.06 | -0.02 | -0.01 | -143.50 | 29.00 | 0.51 | 2018-11-30 |
| 0.07 | 0.01 | -0.02 | -0.03 | -0.03 | 0.00 | -0.05 | 0.00 | -133.50 | 41.00 | 0.51 | 2019-02-28 |
| 0.07 | 0.07 | 0.05 | 0.10 | 0.03 | 0.03 | 0.03 | 0.04 | -154.00 | 52.75 | 0.51 | 2018-07-26 |
| -0.02 | -0.05 | -0.04 | -0.11 | -0.07 | -0.05 | -0.06 | 0.00 | -129.25 | 37.25 | 0.51 | 2019-06-07 |
| 0.01 | 0.09 | 0.00 | 0.00 | 0.01 | -0.01 | 0.01 | 0.04 | -152.50 | 44.75 | 0.51 | 2019-03-09 |
| 0.00 | 0.04 | 0.03 | 0.05 | 0.09 | -0.04 | 0.04 | 0.07 | -133.75 | 47.75 | 0.51 | 2018-05-07 |
| -0.01 | 0.09 | 0.01 | 0.06 | -0.04 | 0.04 | 0.01 | -0.04 | -139.00 | 51.75 | 0.51 | 2018-07-07 |
| -0.07 | 0.08 | 0.01 | 0.13 | -0.10 | 0.03 | -0.06 | -0.19 | -127.00 | 28.75 | 0.51 | 2018-07-03 |
How to visualize it?
Well there are plenty of ways to display data within a netCDF. Here,
we are only presenting one simple way to do it using
ggplot2 and gganimate package.
# first let's calculate the velocity norm
df[, `:=`(
vel_cglo = sqrt(uo_cglo^2 + vo_cglo^2),
vel_oras = sqrt(uo_oras^2 + vo_oras^2),
vel_foam = sqrt(uo_foam^2 + vo_foam^2),
vel_glor = sqrt(uo_glor^2 + vo_glor^2),
month = month(time),
year = year(time)
)]
# average by month
df_summarize <- df[, .(
vel_cglo = mean(vel_cglo, na.rm = T),
vel_oras = mean(vel_oras, na.rm = T),
vel_foam = mean(vel_foam, na.rm = T),
vel_glor = mean(vel_glor, na.rm = T)
),
by = .(
date = paste(year, "-", month),
longitude,
latitude
)
] %>%
.[, time := .GRP, by = .(date)]
# data wrangling
dataPlot <- melt(df_summarize,
id.vars = c("date", "longitude", "latitude", "time"),
variable.name = "model"
)
# let's comput the ouput of all models
res_anim <- ggplot() +
geom_raster(
data = dataPlot,
aes(x = longitude, y = latitude, fill = value),
interpolate = TRUE
) +
geom_sf(
data = spData::world,
col = 1,
fill = "ivory"
) +
coord_sf(xlim = c(-165, -115), ylim = c(25, 55)) +
facet_wrap(. ~ model, ncol = 2) +
theme_jjo() +
theme(legend.position = "bottom") +
labs(x = NULL, y = NULL) +
scale_fill_gradientn(colours = oce::oceColors9A(120)) +
labs(title = "Date: {df_summarize[,unique(date)][frame_time]}") +
transition_time(time) +
ease_aes("linear")
# let's print
animate(res_anim, height = 500, width = 500)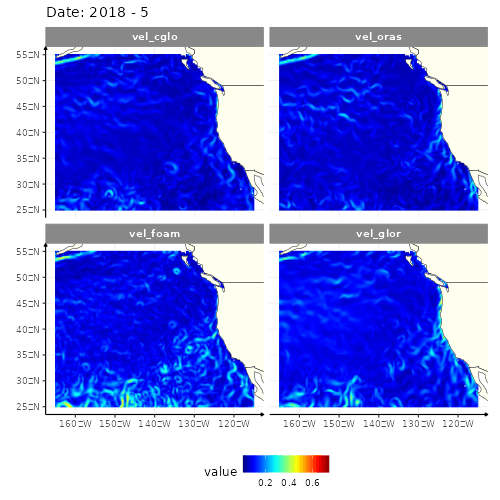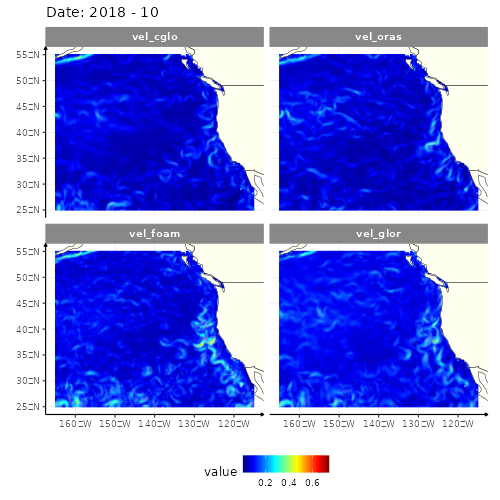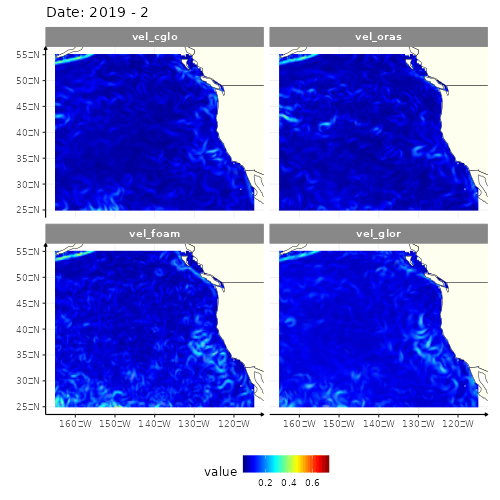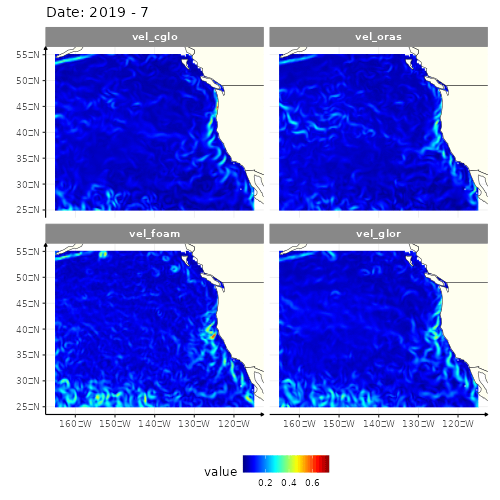
4 models of the current velocity in the North-East part of Pacific ocean
Four different GIFs, since there are four different models to get the
SST: 
