1 Import Data
# load wealingNES package
library(ontodive)
# load dataset
data_nes <- get_data("nes")
data_ses <- get_data("ses")
# merge into one dataset
data_comp <- rbind(
rbindlist(data_nes$year_2018),
rbindlist(data_ses$year_2014),
use.names = T,
fill = T
)
# remove outlier (i.e. dive duration > 5000 s)
data_comp <- data_comp[dduration < 5000, ]
data_comp[, diff_days := c(0, diff(day_departure)), by = .id]
# keep the first trip
data_comp_split <- split(data_comp, by = c(".id"))
data_comp_split_list <- lapply(data_comp_split, function(x) {
# find the rows after 100 day_departure where diff_days > 1
second_trip <- x[day_departure > 100 & diff_days > 1, ]
# if there is a second trip
if (nrow(second_trip) != 0) {
# get the first date of this second trip
date_cut <- second_trip[, min(date)]
# return only the first trip
return(x[date < date_cut, ])
# if no second trip
} else {
# return the full dataset
return(x)
}
})
# rebuilt the dataset
data_comp <- rbindlist(data_comp_split_list)
# rename sp for viz purposes
data_comp %>% .[, sp_rename := fifelse(
sp == "nes",
"Northern elephant seal",
"Southern elephant seal"
)]
# rename divetype for viz purposes
data_comp[, divetype_rename := divetype %>%
word(2) %>%
str_to_title()]
data_comp[divetype_rename == "Foraging",
divetype_rename := "Active Bottom"]
# set up colours
# https://davidmathlogic.com/colorblind/#%23D81B60-%231E88E5-%23FFC107-%23004D40
# https://thenode.biologists.com/data-visualization-with-flying-colors/research/
# https://color.adobe.com/create/color-accessibility
# colours <- c("#e08214", "#8073ac")
colours <- c("#E1BE6A", "#009E73")2 Figures
Figure 1
# datasets
data_fig_sum <- data_comp[!is.na(lat) & .id %in% c(
"ind_2018070",
"ind_140072"
), ] %>%
.[.id == "ind_2018070", .id := "Northern elephant seal: ID 2018070"] %>%
.[.id == "ind_140072", .id := "Southern elephant seal: ID 140072"] %>%
.[, .id := ordered(.id, levels = c(
"Northern elephant seal: ID 2018070",
"Southern elephant seal: ID 140072"
))]
# maxdepth
data_fig_sum_maxdepth <- melt(
data_fig_sum[, .(
date,
maxdepth,
day_departure,
.id
)],
id.vars = c("date", "day_departure", ".id"),
measure.vars = c("maxdepth")
)
# duration
data_fig_sum_dduration <- melt(
data_fig_sum[, .(
date,
dduration,
day_departure,
.id
)],
id.vars = c("date", "day_departure", ".id"),
measure.vars = c("dduration")
)
# driftrate
data_fig_sum_driftrate <- melt(
data_fig_sum[divetype == "2: drift", .(
driftrate = median(driftrate, na.rm = T),
day_departure = first(day_departure)
),
by = .(date = as.Date(date), .id)
],
id.vars = c("date", "day_departure", ".id"),
measure.vars = c("driftrate")
)
# bADL
data_fig_sum_adl <- melt(
data_fig_sum[divetype == "1: foraging",
.(
adl = round(quantile(dduration, 0.95) /
60),
day_departure = first(day_departure)
),
by = .(date = as.Date(date), .id)
],
id.vars = c("date", "day_departure", ".id"),
measure.vars = c("adl")
)
# configure theme (to get gradient https://encycolorpedia.com/)
nes_theme <- ttheme(
colnames.style = colnames_style(
color = "black",
fill = colours[1],
face = "bold"
),
tbody.style = tbody_style(color = "black", fill = c("#e8c883", "#f3deb4"))
)
ses_theme <- ttheme(
colnames.style = colnames_style(
color = "black",
fill = colours[2],
face = "bold"
),
tbody.style = tbody_style(color = "black", fill = c("#74bfa0", "#bbdfce"))
)
# map
trip <- basemap(
shapefiles = "DecimalDegree",
bathymetry = TRUE
) +
geom_path(
data = data_fig_sum,
aes(x = lon, y = lat, group = .id),
linewidth = 2,
colour = "white",
show.legend = F
) +
scale_fill_manual(
name = "Bathymetry (m)",
values = colorRampPalette(c(
"#F7FBFF", "#DEEBF7", "#9ECAE1", "#4292C6", "#08306B"
))(10),
labels =
c(
"0-50",
"50-300",
"300-500",
"500-1000",
"1000-1500",
"1500-2000",
"2000-4000",
"4000-6000",
"6000-10000",
">10000"
)
) +
geom_path(
data = data_fig_sum,
aes(x = lon, y = lat, col = .id),
linewidth = 1.5,
show.legend = F
) +
scale_color_manual(values = colours, guide = "none") +
# annotation_raster(nes_image, -175, -145, 20, 50) +
# annotation_raster(ses_image, 75, 105, -50, -20) +
theme_void() +
theme(legend.position = "top")
# maxdepth
fig_sum_maxdepth <-
ggplot(data_fig_sum_maxdepth, aes(x = day_departure, y = value, col = .id)) +
geom_point(
show.legend = F,
shape = 16,
size = 1,
alpha = 0.5
) +
labs(y = "Maximum depth (m)") +
scale_y_reverse() +
scale_colour_manual(values = colours) +
facet_grid2(. ~ .id,
strip = strip_themed(background_x = elem_list_rect(fill = colours))
) +
theme_jjo() +
theme(
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
strip.background = element_blank(),
strip.text.x = element_blank()
)
# driftrate
fig_sum_dduration <-
ggplot(
data_fig_sum_dduration,
aes(x = day_departure, y = value / 60, col = .id)
) +
geom_point(
show.legend = F,
shape = 16,
size = 1,
alpha = 0.5
) +
labs(y = "Dive duration (min)") +
scale_colour_manual(values = colours) +
facet_grid2(. ~ .id,
strip = strip_themed(background_x = elem_list_rect(fill = colours))
) +
theme_jjo() +
theme(
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
strip.background = element_blank(),
strip.text.x = element_blank()
)
# driftrate
fig_sum_driftrate <-
ggplot(
data_fig_sum_driftrate,
aes(x = day_departure, y = value, col = .id)
) +
geom_point(show.legend = F) +
geom_hline(
yintercept = 0,
linetype = "dashed",
size = 1
) +
labs(
y = expression(paste("Daily median drift rate (m.", s^-1, ")")),
x = "Number of days since departure"
) +
scale_colour_manual(values = colours) +
facet_grid2(. ~ .id,
strip = strip_themed(background_x = elem_list_rect(fill = colours))
) +
theme_jjo() +
theme(
strip.background = element_blank(),
strip.text.x = element_blank()
)
# summary table for nes
table_sum_nes <- data.table::transpose(data_fig_sum[sp == "nes", .(
"# of days recorded" = prettyNum(
uniqueN(as.Date(date)),
big.mark = ",",
scientific = FALSE
),
"Recorder settings" = "All dives",
"# of dives recorded" = prettyNum(.N, big.mark = ",", scientific = FALSE),
"Median max depth" = paste(prettyNum(
round(quantile(maxdepth, 0.5), 1),
big.mark = ",",
scientific = FALSE
), "m"),
"Median dive duration" = paste(prettyNum(
round(quantile(dduration, 0.5) / 60, 1),
big.mark = ",",
scientific = FALSE
), "min")
), by = .("Seal ID" = .id)], keep.names = " ", make.names = 1)
# summary table for ses
table_sum_ses <- data.table::transpose(data_fig_sum[sp == "ses", .(
"# of days recorded" = prettyNum(
uniqueN(as.Date(date)),
big.mark = ",",
scientific = FALSE
),
"Recorder settings" = "1 dive every ~2.25 h",
"# of dives recorded" = prettyNum(.N, big.mark = ",", scientific = FALSE),
"Median max depth" = paste(prettyNum(
round(quantile(maxdepth, 0.5), 1),
big.mark = ",",
scientific = FALSE
), "m"),
"Median dive duration" = paste(prettyNum(
round(quantile(dduration, 0.5) / 60, 1),
big.mark = ",",
scientific = FALSE
), "min")
), by = .("Seal ID" = .id)], keep.names = " ", make.names = 1)
# format tables
table_sum_nes <- tableGrob(table_sum_nes,
rows = NULL,
cols = NULL,
theme = nes_theme
)
header_sum_nes <- tableGrob(mtcars[1, 1],
rows = NULL,
cols = c(data_fig_sum[sp == "nes", unique(.id)]),
theme = nes_theme
)
table_sum_nes <- gtable_combine(header_sum_nes[1, ], table_sum_nes, along = 2)
table_sum_nes$layout[1:2, c("r")] <- 2
table_sum_nes$widths <- unit(c(0.6, 0.4), "npc")
table_sum_ses <- tableGrob(table_sum_ses,
rows = NULL,
cols = NULL,
theme = ses_theme
)
header_sum_ses <- tableGrob(mtcars[1, 1],
rows = NULL,
cols = c(data_fig_sum[sp == "ses", unique(.id)]),
theme = ses_theme
)
table_sum_ses <- gtable_combine(header_sum_ses[1, ], table_sum_ses, along = 2)
table_sum_ses$layout[1:2, c("r")] <- 2
table_sum_ses$widths <- unit(c(0.6, 0.4), "npc")
layout <- "
AAAADDD
AAAADDD
AAAADDD
AAAADDD
AAAADDD
AAAADDD
AAAAEEE
AAAAEEE
AAAAEEE
AAAAEEE
AAAAEEE
BBBBEEE
BBBBFFF
CCCCFFF
CCCCFFF
CCCCFFF
CCCCFFF
CCCCFFF
"
# plot_to_save <-
trip +
guide_area() +
(as_ggplot(table_sum_nes) + as_ggplot(table_sum_ses)) +
fig_sum_maxdepth +
fig_sum_dduration +
fig_sum_driftrate +
plot_annotation(tag_levels = list(c("(A)", "(B)", "", "(C)", "(D)", "(E)"))) +
# layout
plot_layout(design = layout, guides = "collect") &
# annotation in bold
theme(plot.tag = element_text(face = "bold"))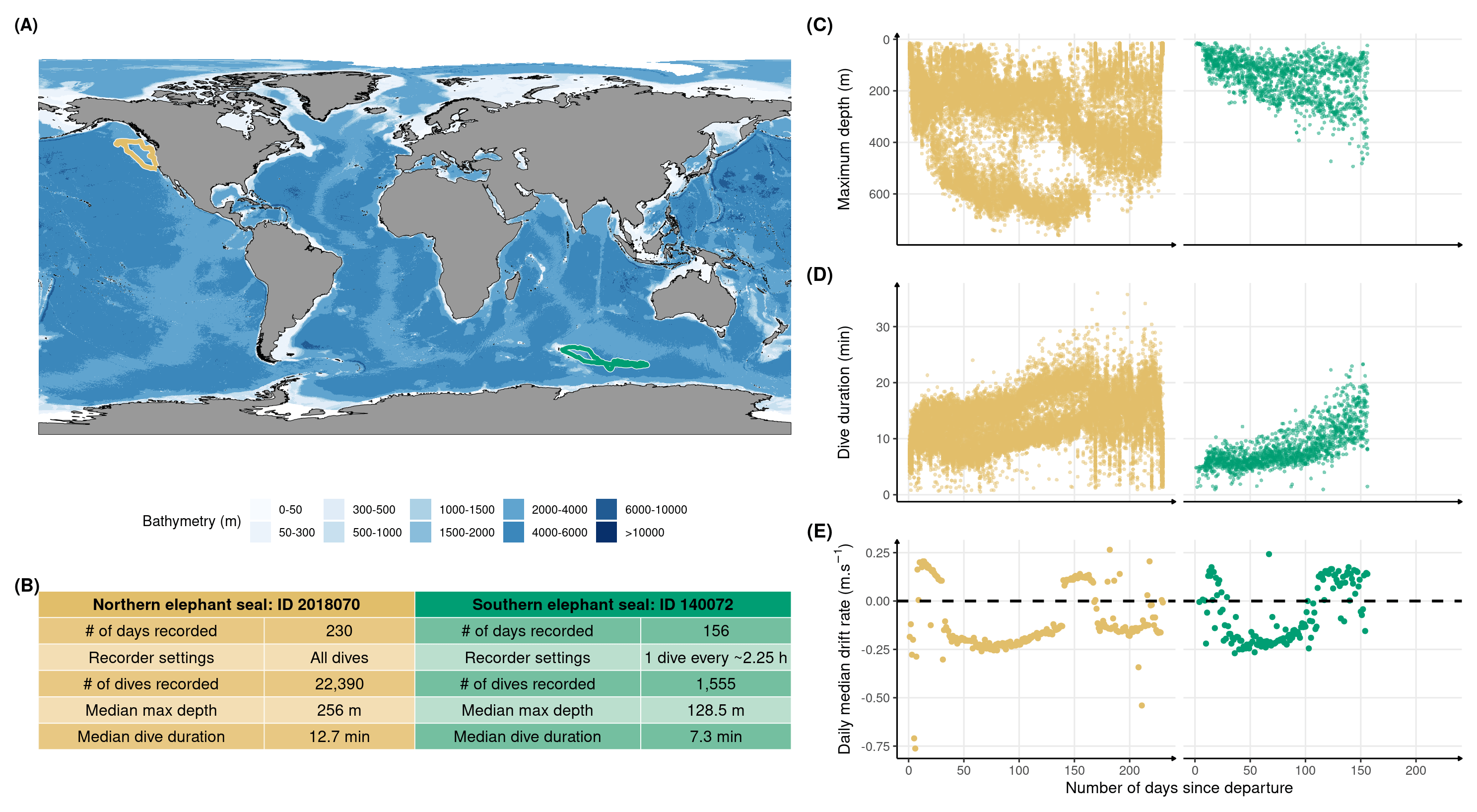
Figure 2.1: Illustrative data from one representative weanling northern elephant seal (id: 2018070) and one southern elephant seal (id: 130072) for comparison. The panels show for each of these two representative seals: (A) the migration routes during their first trip to sea from Año Nuevo, California, United States of America, and Kerguelen Island, France, respectively in gold and green; (B) a summary of tag setup and dive characteristics; the development of (C) maximum diving depth, (D) dive duration, and (E) daily median drift rate. Data show clear improvement of diving metrics for both species, with northern elephant seals exhibiting accelerated development in both diving duration and depth.
# export (height 785; width 1494)
# ggsave("test.png", width = 180, height = 90, units = "mm", dpi = 300, scale = 2.5)
# save the plot
# ggsave("figure_1.png",
# plot_to_save,
# width = 180,
# height = 90,
# units = "mm",
# dpi = 300,
# scale = 2.5)Figure 2
# initial plots
fig_maxdepth_ini <- plot_comp(
data_comp,
"maxdepth",
group_to_compare = "sp_rename",
nb_days = 200,
cols = "divetype_rename",
ribbon = T,
point = F,
colours = colours,
linetype_ribbon = 0,
individual = FALSE,
# method = "GCV.Cp",
scales = "free_y"
)
fig_dduration_ini <- plot_comp(
copy(data_comp)[, dduration_min := dduration / 60],
"dduration_min",
group_to_compare = "sp_rename",
nb_days = 200,
cols = "divetype_rename",
ribbon = T,
point = F,
colours = colours,
linetype_ribbon = 0,
individual = FALSE,
# method = "GCV.Cp",
scales = "free_y"
)
# get limits
maxdepth_limits <- ggplot_build(fig_maxdepth_ini)$layout$panel_params[[1]]$y.range
dduration_limits <- ggplot_build(fig_dduration_ini)$layout$panel_params[[1]]$y.range
# update initial plots
fig_maxdepth <- fig_maxdepth_ini +
labs(
y = "Maximum depth (m)",
colour = "Elephant seals",
fill = "Elephant seals"
) +
coord_cartesian(ylim = rev(maxdepth_limits)) +
scale_colour_manual(
values = colours,
labels = data_comp[, sort(unique(sp_rename))] %>%
word(1) %>%
str_to_title()
) +
scale_fill_manual(
values = colours,
labels = data_comp[, sort(unique(sp_rename))] %>%
word(1) %>%
str_to_title()
) +
theme_jjo() +
theme(
legend.position = "top",
axis.title.x = element_blank(),
axis.text.x = element_blank(),
axis.line.x = element_blank(),
axis.ticks.x = element_blank(),
strip.text = element_text(colour = "grey20"),
axis.line.y = element_line(color = "black")
)
fig_dduration <- fig_dduration_ini +
labs(
x = "Number of days since departure",
y = "Dive duration (min)"
) +
coord_cartesian(ylim = dduration_limits) +
theme_jjo() +
theme(
legend.position = "none",
strip.text.x = element_blank(),
axis.line.y = element_line(color = "black")
)
# density plots
fig_dens_maxdepth <-
ggplot(data_comp[day_departure <= 200, ], aes(y = maxdepth, fill = sp)) +
geom_density(show.legend = F, col = "black", alpha = 0.4, size = 0.3) +
coord_cartesian(ylim = rev(maxdepth_limits)) +
scale_fill_manual(values = colours) +
theme_void()
fig_dens_dduration <-
ggplot(data_comp[day_departure <= 200, ], aes(y = dduration / 60, fill = sp)) +
geom_density(show.legend = F, col = "black", alpha = 0.4, size = 0.3) +
coord_cartesian(ylim = dduration_limits) +
scale_fill_manual(values = colours) +
theme_void()
# plot_to_save <-
((fig_maxdepth | fig_dens_maxdepth) + plot_layout(ncol = 2, widths = c(5, 1))) /
((fig_dduration | fig_dens_dduration) + plot_layout(ncol = 2, widths = c(5, 1)))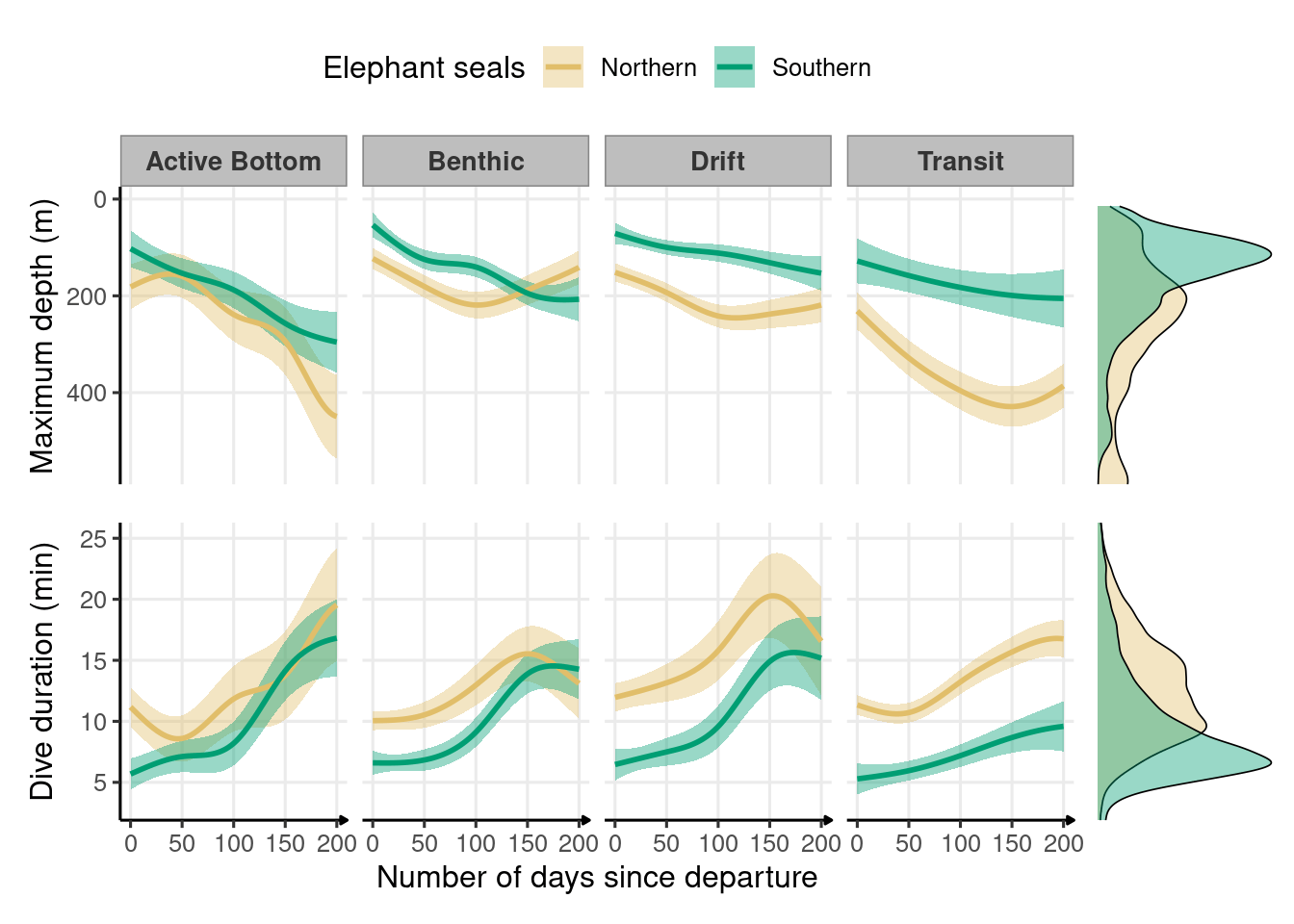
Figure 2.2: Development of depth and duration across dive types throughout the first trip to sea in northern (n = 4) and southern elephant seals (n = 9) estimated from generalized additive models. The solid lines represent the means, and the shaded areas represent the 95% confidence intervals. Marginal density plots represent the spread of data across all dive types for each species.
# # save the plot
# ggsave(filename = "figure_2.png",
# plot = plot_to_save,
# width = 9,
# height = 6)
# Mann Whitney / Wilcoxon rank sum test on dive depth
tidy(wilcox.test(
data_comp[day_departure <= 200 & sp == "nes", maxdepth],
data_comp[day_departure <= 200 & sp == "ses", maxdepth]
)) %>%
gt() %>%
tab_header(title = "Mann Whitney / Wilcoxon rank sum test on dive depth")| Mann Whitney / Wilcoxon rank sum test on dive depth | |||
| statistic | p.value | method | alternative |
|---|---|---|---|
| 902643075 | 0 | Wilcoxon rank sum test with continuity correction | two.sided |
# Mann Whitney / Wilcoxon rank sum test on dive duration
tidy(wilcox.test(
data_comp[day_departure <= 200 & sp == "nes", dduration],
data_comp[day_departure <= 200 & sp == "ses", dduration]
)) %>%
gt() %>%
tab_header(title = "Mann Whitney / Wilcoxon rank sum test on dive duration")| Mann Whitney / Wilcoxon rank sum test on dive duration | |||
| statistic | p.value | method | alternative |
|---|---|---|---|
| 897660292 | 0 | Wilcoxon rank sum test with continuity correction | two.sided |
Figure 3
# (only for .id with location data, and so phase information)
prop_dive_id_phase_divetype_sp <- data_comp[
!is.na(lat),
table(divetype_rename, sp, sp_rename, phase, .id)
] %>%
# the calculate the proportion of dive in each divetype, per sp and phases
prop.table(., c(".id")) %>%
# convert into a data.table
as.data.table(.)
# merge this table to add the number of dives, per divetype, phase, sp, .id
prop_dive_id_phase_divetype_sp <-
merge(
prop_dive_id_phase_divetype_sp,
data_comp[!is.na(lat),
.(nb_dives_divetype = uniqueN(divenumber)),
by = .(sp, sp_rename, .id, divetype_rename, phase)
] %>%
.[, nb_dives := sum(nb_dives_divetype),
by = .(.id)
] %>%
.[],
by = c("sp_rename", "sp", ".id", "divetype_rename", "phase"),
all.y = T
)
# calculate the right proportions
dataPlot <- prop_dive_id_phase_divetype_sp %>%
.[, .(
N = wtd.mean(N, nb_dives),
# its equivalent of using only the number of dives and not the percentage
# N == N_v2
N_v2 = sum(nb_dives_divetype) / sum(nb_dives),
N_sd = sqrt(wtd.var(N, nb_dives))
),
by = .(sp_rename, sp, divetype_rename, phase)
]
# p_value calculation for nes
df_p_val_nes <- data_comp[sp == "nes" & !is.na(lat),
.(nb_divetype = .N),
by = .(divetype_rename, phase)
] %>%
.[, nb_dive := sum(nb_divetype)] %>%
# perform by divetype
rstatix::group_by(divetype_rename) %>%
# a prop.test test
summarise(rstatix::prop_test(x = nb_divetype, n = nb_dive, correct = F)) %>%
# then adjust the p_value for multiple test
rstatix::adjust_pvalue(p.col = "p", method = "bonferroni") %>%
# update p.adj
rstatix::add_significance(p.col = "p.adj") %>%
# sort
arrange(divetype_rename)
# dataset for nes
dataPlot_nes <- copy(dataPlot)[sp == "nes", N := -(1 * N)] %>%
.[sp == "nes"]
# plot
fig_nes_prop <-
ggplot(
dataPlot_nes,
aes(x = divetype_rename, y = N, fill = phase)
) +
geom_bar(
stat = "identity",
position = "dodge",
color = "grey30"
) +
scale_y_continuous(
labels = function(x) {
percent(abs(x), 1)
}
) +
geom_errorbar(aes(ymin = N - N_sd, ymax = N),
width = .2,
position = position_dodge(.9)
) +
coord_flip(ylim = c(-c(round((dataPlot[, max(N + N_sd)] + 0.03) * 100) / 100, 0))) +
facet_grid(. ~ sp_rename, scales = "free_x") +
theme_jjo() +
# day first, and night
scale_fill_manual(
values = c("white", "grey"),
labels = c("Day-time", "Night-time")
) +
# add stat
geom_signif(
y_position = dataPlot_nes[, .(position = min(N - N_sd)), divetype_rename]$position +
dataPlot_nes[, max(N + N_sd)] * 0.5,
xmin = seq(0, dataPlot_nes[, uniqueN(divetype_rename)] - 1) + 0.8,
xmax = seq(1, dataPlot_nes[, uniqueN(divetype_rename)]) + 0.2,
# replace **** by *** (more standard)
annotation = fifelse(
df_p_val_nes$p.adj.signif == "****",
"***",
df_p_val_nes$p.adj.signif
),
tip_length = 0,
vjust = -0.6,
angle = 90
) +
labs(fill = "Time of day") +
theme(
legend.position = "top",
axis.line.y = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line = element_line(arrow = arrow(
length = unit(0.2, "lines"),
type = "closed",
ends = "first"
)),
strip.background = element_rect(fill = colours[1]),
strip.text = element_text(colour = "grey20")
)
# p_value calculation for ses
df_p_val_ses <- data_comp[sp == "ses" & !is.na(lat),
.(nb_divetype = .N),
by = .(divetype_rename, phase)
] %>%
.[, nb_dive := sum(nb_divetype)] %>%
# perform by divetype
rstatix::group_by(divetype_rename) %>%
# a prop.test test
summarise(rstatix::prop_test(x = nb_divetype, n = nb_dive, correct = F)) %>%
# then adjust the p_value for multiple test
rstatix::adjust_pvalue(p.col = "p", method = "bonferroni") %>%
# update p.adj
rstatix::add_significance(p.col = "p.adj") %>%
# sort
arrange(divetype_rename)
# dataset for ses
dataPlot_ses <- copy(dataPlot)[sp == "ses", ] %>%
.[sp == "ses"]
# plot
fig_ses_prop <-
ggplot(dataPlot_ses, aes(x = divetype_rename, y = N, fill = phase)) +
geom_bar(
stat = "identity",
position = "dodge",
color = "grey30"
) +
scale_y_continuous(
labels = function(x) {
percent(abs(x), 1)
}
) +
geom_errorbar(aes(ymin = N, ymax = N + N_sd),
width = .2,
position = position_dodge(.9)
) +
coord_flip(ylim = c(0, round((dataPlot[, max(N + N_sd)] + 0.03) * 100) / 100)) +
facet_grid(. ~ sp_rename, scales = "free_x") +
theme_jjo() +
# day first, and night
scale_fill_manual(
values = c("white", "grey"),
labels = c("Day-time", "Night-time")
) +
# add stat
geom_signif(
y_position = dataPlot_ses[, .(position = max(N + N_sd)), divetype_rename]$position +
dataPlot_ses[, min(N + N_sd)] * 0.5,
xmin = seq(0, dataPlot_ses[, uniqueN(divetype_rename)] - 1) + 0.8,
xmax = seq(1, dataPlot_ses[, uniqueN(divetype_rename)]) + 0.2,
# replace **** by *** (more standard)
annotation = fifelse(
df_p_val_ses$p.adj.signif == "****",
"***",
df_p_val_ses$p.adj.signif
),
tip_length = 0
) +
labs(fill = "Time of day") +
theme(
legend.position = "none",
axis.line.y = element_blank(),
axis.title.y = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
axis.line.x = element_line(arrow = arrow(
length = unit(0.2, "lines"), type = "closed"
)),
strip.background = element_rect(fill = colours[2]),
strip.text = element_text(colour = "grey20")
)
df_text <- data.table(
x = rep(0, data_comp[, uniqueN(divetype_rename)]),
label_to_order = data_comp[, sort(unique(divetype_rename))],
divetype = copy(data_comp)[divetype_rename == "Active Bottom",
divetype_rename := "Active Btt."] %>%
.[, sort(unique(divetype_rename))],
percentage = round(as.vector(prop.table(data_comp[
!is.na(lat),
table(divetype_rename)
])) * 100, 1)
) %>%
.[, label_to_display := paste0(divetype, "\n(", percentage, " %)")]
fig_text <-
ggplot(df_text, aes(x = x, y = label_to_order, label = label_to_display)) +
geom_text() +
theme_void()
fig_label_1 <-
ggplot(data.frame(l = "Percentage of dives", x = 1, y = 1)) +
geom_text(aes(x, y, label = l)) +
theme_void() +
coord_cartesian(clip = "off")
((fig_nes_prop | fig_text | fig_ses_prop) +
plot_layout(
widths = c(7, 2, 7),
guides = "collect"
) &
theme(legend.position = "top")) / fig_label_1 +
plot_layout(heights = c(6, 1))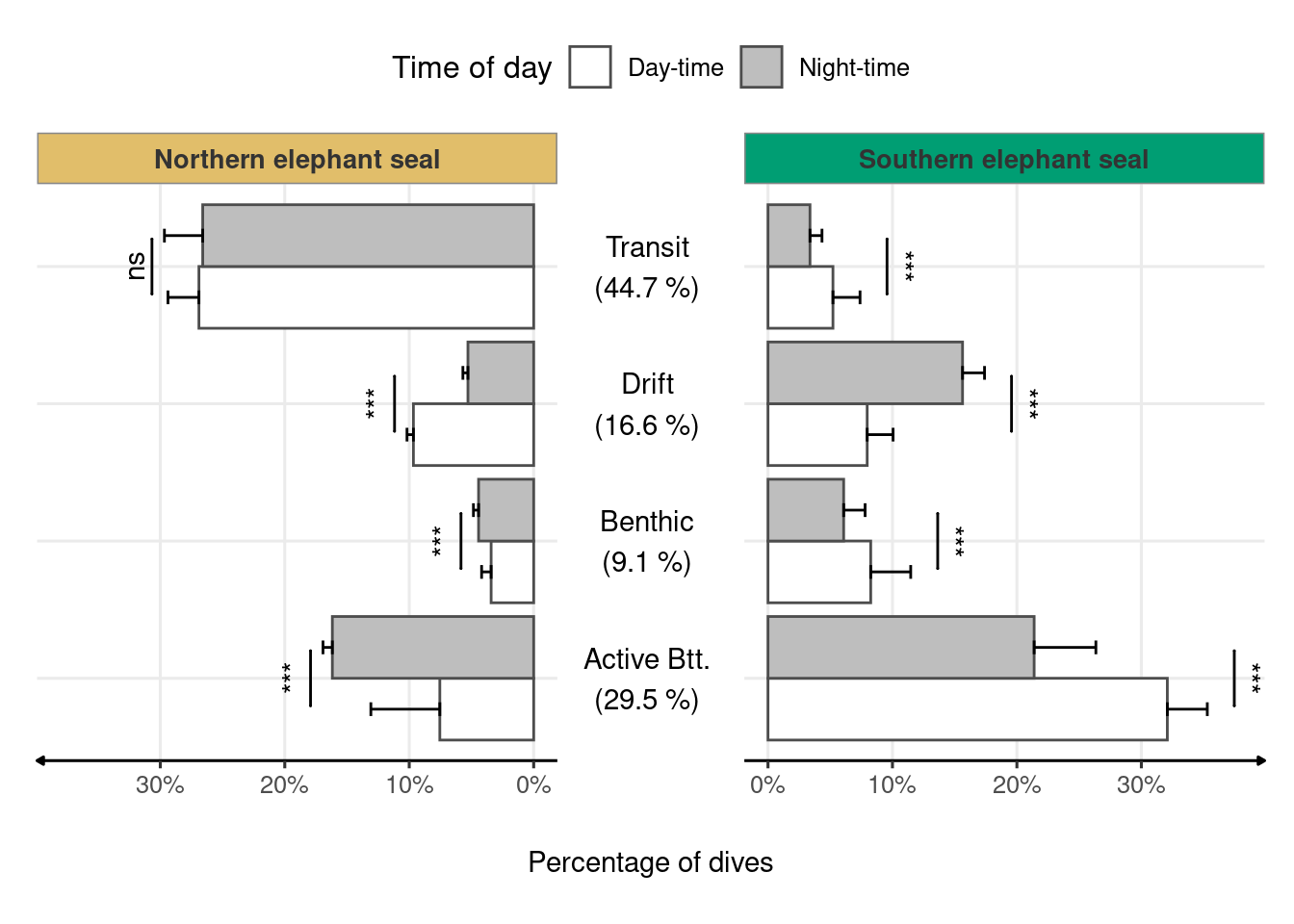
Figure 2.3: Frequency of dive types across time of day and species. Species-wide statistical tests were based on averages of each individual’s dive type proportion weighted by the total number of dives. Percentages in the middle panel represent the frequency of each dive type across species and time of day. Asterisks (*) indicate a significant difference (P-value < 0.0001) between day-time and night-time dive proportions within each species (two-sample z-test).
Figure 4
# calculate the median of driftrate for each day
median_driftrate <- data_comp[divetype == "2: drift",
.(driftrate = quantile(driftrate, 0.5)),
by = .(day_departure, .id, sp)
] %>%
.[, sp := fifelse(sp == "nes", "Northern", "Southern")]
# initial plots
fig_driftrate_ini <- plot_comp(
median_driftrate,
"driftrate",
group_to_compare = "sp",
nb_days = 200,
ribbon = T,
linetype_ribbon = 0,
point = F,
colours = colours
)
# get limits
driftrate_limits <- ggplot_build(fig_driftrate_ini)$layout$panel_params[[1]]$y.range
# update initial plots
fig_driftrate <- fig_driftrate_ini +
labs(
y = expression(paste("Daily median drift rate (m.", s^-1, ")")),
x = "Number of days since departure",
colour = "Elephant seal",
fill = "Elephant seal"
) +
geom_hline(
yintercept = 0,
linetype = 2,
size = 1,
col = "black"
) +
coord_cartesian(ylim = driftrate_limits) +
theme_jjo() +
theme(
legend.position = "top"
)
# density plots
fig_dens_driftrate <-
ggplot(
data_comp[divetype == "2: drift" & day_departure <= 200, ],
aes(y = driftrate, fill = sp)
) +
geom_density(show.legend = F, col = "black", alpha = 0.4, size = 0.3) +
coord_cartesian(ylim = driftrate_limits) +
scale_fill_manual(values = colours) +
theme_void()
(fig_driftrate | fig_dens_driftrate) + plot_layout(widths = c(5, 1))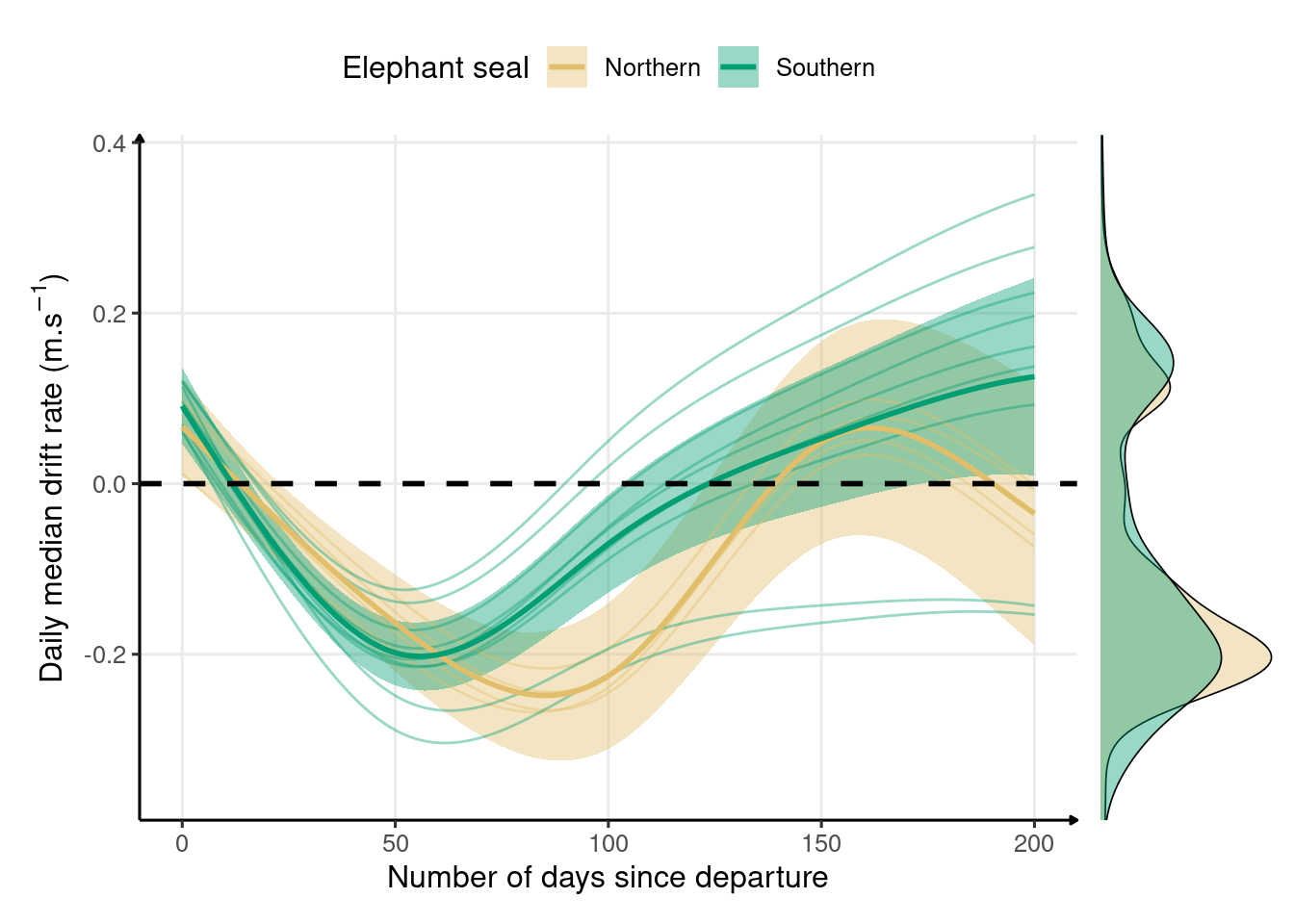
Figure 2.4: Changes in median drift rate across the first trip to sea in northern (n = 4) and southern (n = 9) elephant seals estimated from a generalized additive model. The bold solid lines represent the mean species-level responses while the thin lines represent individual-level responses. The shaded areas represent the 95% confidence interval, and the black dashed line indicates neutral buoyancy. Marginal density plots indicate the data spread across the entire migration for each species.
Appendix
# initial plots
max_depth_all_res <- plot_comp(
data_comp %>%
.[, sp := fifelse(sp == "nes", "Northern", "Southern")],
"maxdepth",
group_to_compare = "sp",
nb_days = 200,
ribbon = T,
linetype_ribbon = 0,
point = F,
colours = colours,
export_data_model = T
)
max_depth_all_ini <- max_depth_all_res[[1]]
max_depth_all_data <- max_depth_all_res[[2]]
# https://stackoverflow.com/questions/70420256/how-to-draw-a-horizontal-line-and-a-vertical-line-that-cross-at-the-intersection
draw_guides <- function(x, y) {
list(
geom_segment(aes(x = -Inf, xend = x, y = y, yend = y), linetype = "dashed"),
geom_segment(aes(x = x, xend = x, y = y, yend = -Inf), linetype = "dashed")
)
}
# get limits
max_depth_all_limits <- ggplot_build(max_depth_all_ini)$layout$panel_params[[1]]$y.range
# update initial plots
max_depth_all <- max_depth_all_ini +
labs(
y = "Maximum depth (m)",
x = "Number of days since departure",
colour = "Elephant seal",
fill = "Elephant seal"
) +
coord_cartesian(ylim = rev(max_depth_all_limits)) +
scale_fill_manual(values = colours) +
geom_point(data = data.table(x = c(30, 160), y = c(240, 240)), aes(x, y)) +
apply(data.table(x = c(30, 160), y = c(240, 240)), 1, function(dt) {
draw_guides(dt[1], dt[2])
}) +
scale_x_continuous(position = "top") +
theme_jjo() +
theme(
legend.position = "bottom",
axis.line.y = element_line(color = "black", arrow = arrow(
length = unit(0.2, "lines"),
type = "closed",
ends = "first"
)),
)
# density plots
fig_dens_max_depth_all <-
ggplot(
data_comp[day_departure <= 200, ],
aes(y = maxdepth, fill = sp)
) +
geom_density(show.legend = F, col = "black", alpha = 0.4, linewidth = 0.3) +
coord_cartesian(ylim = rev(max_depth_all_limits)) +
scale_fill_manual(values = colours) +
theme_void()
# plot
# plot_to_save <-
(max_depth_all | fig_dens_max_depth_all) + plot_layout(widths = c(5, 1))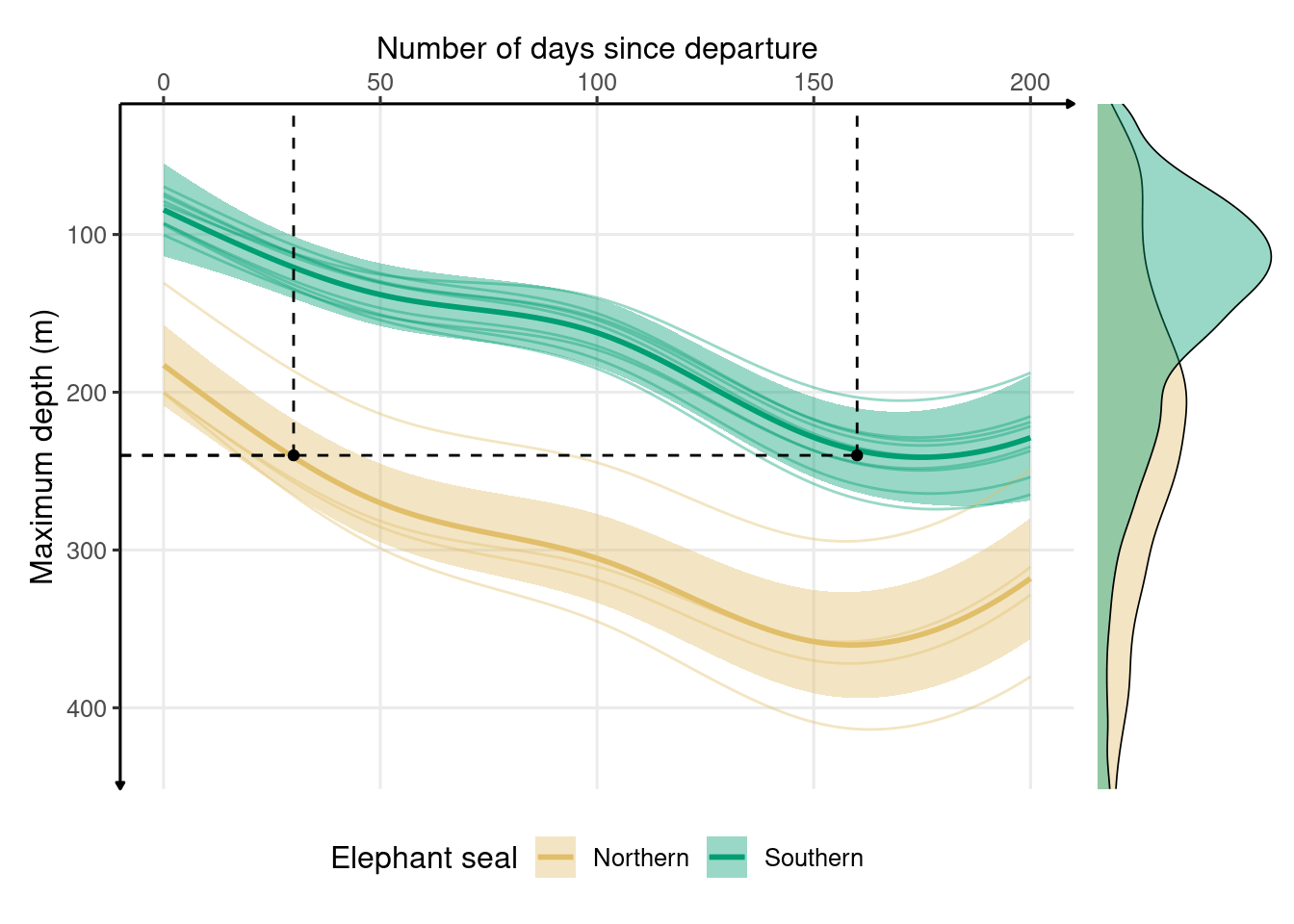
Figure 2.5: Development of depth throughout the first trip to sea in northern (n = 4) and southern elephant seals (n = 9) estimated from generalized additive models. The solid lines represent the means, and the shaded areas represent the 95% confidence intervals. Marginal density plots represent the spread of data for each species. Depth of 260 meters is reached in 30 days for northern elephant seals and 160 for southern elephant seals.
# # to save
# ggsave("figure_supp_1.png",
# plot_to_save,
# width = 6,
# height = 4
# )
# initial plots
dive_duration_all_res <- plot_comp(
copy(data_comp)[, dduration := dduration / 60],
"dduration",
group_to_compare = "sp",
nb_days = 200,
ribbon = T,
linetype_ribbon = 0,
point = F,
colours = colours,
export_data_model = T
)
dive_duration_all_ini <- dive_duration_all_res[[1]]
dive_duration_all_data <- dive_duration_all_res[[2]]
# get limits
dive_duration_all_limits <- ggplot_build(dive_duration_all_ini)$layout$panel_params[[1]]$y.range
# update initial plots
dive_duration_all <- dive_duration_all_ini +
labs(
y = "Dive duration (min)",
x = "Number of days since departure",
colour = "Elephant seal",
fill = "Elephant seal"
) +
geom_point(data = data.table(x = c(0, 125), y = c(11.15, 11.15)), aes(x, y)) +
apply(data.table(x = c(0, 125), y = c(11.15, 11.15)), 1, function(dt) {
draw_guides(dt[1], dt[2])
}) +
coord_cartesian(ylim = dive_duration_all_limits) +
scale_fill_manual(values = colours) +
theme_jjo() +
theme(
legend.position = "top"
)
# density plots
fig_dens_dive_duration_all <-
ggplot(
data_comp[day_departure <= 200, ],
aes(y = dduration / 60, fill = sp)
) +
geom_density(show.legend = F, col = "black", alpha = 0.4, linewidth = 0.3) +
coord_cartesian(ylim = dive_duration_all_limits) +
scale_fill_manual(values = colours) +
theme_void()
# # plot
# plot_to_save <-
(dive_duration_all | fig_dens_dive_duration_all) +
plot_layout(widths = c(5, 1))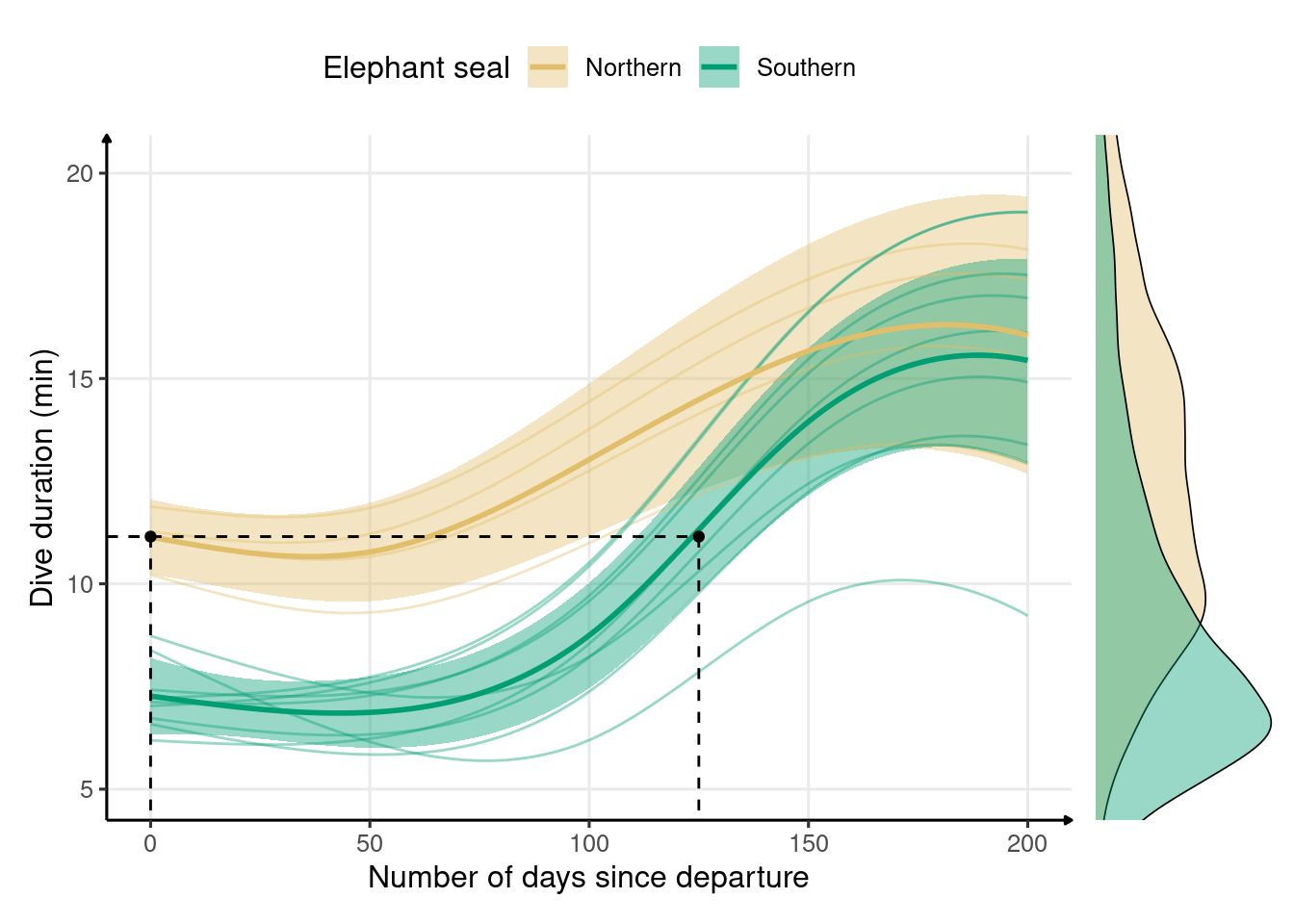
Figure 2.6: Development of dive duration throughout the first trip to sea in northern (n = 4) and southern elephant seals (n = 9) estimated from generalized additive models. The solid lines represent the means, and the shaded areas represent the 95% confidence intervals. Marginal density plots represent the spread of data for each species. Dive duration of 11.15 min is reached on the first day of northern elephant seals’ trip to sea and in 125 days for southern elephant seals.
# # to save
# ggsave("figure_supp_2.png",
# plot_to_save,
# width = 6,
# height = 4)3 Tables
Table 1
I thought about doing a table that provides summary information and gives a rapid overview of the dataset.
# based on
# https://themockup.blog/posts/2020-10-31-embedding-custom-features-in-gt-tables/
gt_ggplot_driftrate <- function(table_data, plot_col, data_col, plot_fun, ...) {
# save the data extract ahead of time
# to be used in our anonymous function below
data_in <- purrr::pluck(table_data, "_data", data_col)
# retrieve min max
range_x <- rbindlist(data_in)[, range(day_departure)]
range_y <- c(-0.35, 0.15)
# draw plot
text_transform(
table_data,
# note the use of {{}} here - this is tidy eval
# that allows you to indicate specific columns
locations = cells_body(columns = c({{ plot_col }})),
fn = function(x) {
# build the plot
plot <- lapply(data_in, function(x) {
# build the plot
ggplot(x) +
# for color https://davidmathlogic.com/colorblind/#%23D81B60-%231E88E5-%23FFC107-%23004D40
geom_area(
aes(x = day_departure, y = ifelse(driftrate < 0,
driftrate, 0
)),
fill = "#5D3A9B", alpha = 0.5
) +
# for color https://davidmathlogic.com/colorblind/#%23D81B60-%231E88E5-%23FFC107-%23004D40
geom_area(
aes(x = day_departure, y = ifelse(driftrate > 0,
driftrate, 0
)),
fill = "#E66100", alpha = 0.5
) +
geom_segment(
aes(
x = 0,
y = 0,
xend = max(day_departure) + 10,
yend = 0
),
size = 7,
colour = "grey30",
arrow = arrow(
type = "closed",
length = unit(0.2, units = "npc")
)
) +
coord_cartesian(xlim = range_x, ylim = range_y) +
theme_void() +
theme(
panel.grid = element_blank(),
panel.border = element_blank(),
)
})
# draw for every row
lapply(plot, ggplot_image, aspect_ratio = 5, height = 25)
}
)
}
gt_ggplot_sparkline <- function(table_data, plot_col, data_col, plot_fun, ...) {
# save the data extract ahead of time
# to be used in our anonymous function below
data_in <- purrr::pluck(table_data, "_data", data_col)
# colnames
col_names <- colnames(rbindlist(data_in))
# interest variable
var_interest <- setdiff(col_names, "day_departure")
# retrieve min max
range_x <- rbindlist(data_in)[, range(day_departure)]
range_y <- rbindlist(data_in)[, range(get(var_interest))]
# draw plot
text_transform(
table_data,
# note the use of {{}} here - this is tidy eval
# that allows you to indicate specific columns
locations = cells_body(columns = c({{ plot_col }})),
fn = function(x) {
# build the plot
plot <- lapply(data_in, function(x) {
# build the plot
ggplot(x, aes(x = day_departure, y = get(var_interest))) +
geom_path(size = 6, color = "grey60") +
geom_smooth(
size = 10,
linetype = "dashed",
colour = "black",
method = "lm",
se = FALSE,
na.rm = TRUE
) +
coord_cartesian(xlim = range_x, ylim = range_y) +
theme_void() +
theme(
panel.grid = element_blank(),
panel.border = element_blank()
)
})
# draw for every row
lapply(plot, ggplot_image, aspect_ratio = 4, height = 26)
}
)
}
# summary_table_es <-
data_comp[, travel_distance := distGeo(
matrix(c(lon, lat), ncol = 2),
matrix(c(shift(lon), shift(lat)), ncol = 2)
),
by = .id
] %>%
as_tibble() %>%
mutate(id_rename = word(.id, 2, sep = "_")) %>%
group_by(sp_rename, id_rename) %>%
summarise(
N = prettyNum(n(),
big.mark = ",",
scientific = FALSE
),
nb_days = round(as.numeric(difftime(
last(date), first(date),
units = "days"
)), 1),
travel_distance = prettyNum(
sum(travel_distance, na.rm = T) / 1000,
digits = 1,
big.mark = ",",
scientific = FALSE
),
Maxdepth_mean = round(mean(maxdepth), 1),
Maxdepth_plus_minus = "±",
Maxdepth_sd = round(sd(maxdepth), 1),
Dduration_mean = round(mean(dduration) / 60, 1),
Dduration_plus_minus = "±",
Dduration_sd = round(sd(maxdepth) / 60, 1),
.groups = "drop"
) %>%
# replace travel_distance = 0 by NA
mutate(travel_distance = na_if(travel_distance, "0")) %>%
# add driftrate from drift dives
left_join(
.,
data_comp %>%
as_tibble() %>%
mutate(id_rename = word(.id, 2, sep = "_")) %>%
group_by(sp_rename, id_rename, day_departure) %>%
filter(divetype == "2: drift") %>%
summarise(
driftrate = as.numeric(quantile(
driftrate, 0.5,
na.rm = T
)),
.groups = "drop"
) %>%
group_by(sp_rename, id_rename) %>%
summarise(
sparkline_driftrate = list(
data.frame(
driftrate = driftrate,
day_departure = day_departure
)
),
.groups = "drop"
),
by = c("sp_rename", "id_rename")
) %>%
# add quantile 95 of dduration and max_depth
left_join(
.,
data_comp %>%
as_tibble() %>%
mutate(id_rename = word(.id, 2, sep = "_")) %>%
group_by(sp_rename, id_rename, day_departure) %>%
summarise(
maxdepth = as.numeric(quantile(maxdepth, 0.95, na.rm = T)),
dduration = as.numeric(quantile(dduration, 0.95, na.rm = T)),
.groups = "drop"
) %>%
group_by(sp_rename, id_rename) %>%
summarise(
sparkline_qt_dduration = list(
data.frame(
dduration = dduration,
day_departure = day_departure
)
),
sparkline_qt_maxdepth = list(
data.frame(
maxdepth = -maxdepth,
day_departure = day_departure
)
),
.groups = "drop"
),
by = c("sp_rename", "id_rename")
) %>%
# add percentage
left_join(
.,
data_comp %>%
as_tibble() %>%
mutate(id_rename = word(.id, 2, sep = "_")) %>%
group_by(sp_rename, id_rename, divetype) %>%
summarise(
n = n(),
.groups = "drop"
) %>%
group_by(sp_rename, id_rename) %>%
mutate(
divetype_perc = round(n * 100 / sum(n)),
divetype,
.groups = "drop"
) %>%
arrange(divetype) %>%
group_by(sp_rename, id_rename) %>%
summarise(divetype_perc = list(divetype_perc), .groups = "drop"),
by = c("sp_rename", "id_rename")
) %>%
# add wean mass based on Weanling Dive Metada.xlsx
left_join(
.,
data.table(
id_rename = c(
"2018070",
"2018072",
"2018074",
"2018080",
"140059",
"140060",
"140062",
"140063",
"140068",
"140069",
"140072",
"140073",
"140075"
),
sp_rename = c(
rep("Northern elephant seal", 4),
rep("Southern elephant seal", 9)
),
weanmass = c(132, 138, 119, 142, 118, 112, 85, NA, 125, NA, NA, 102, NA)
),
by = c("sp_rename", "id_rename")
) %>%
# add wean length based on JoffreyMeasurements 2023_02_01.xlsx and
# metadata_pups20142015.xlsx
left_join(
.,
data.table(
id_rename = c(
"2018070",
"2018072",
"2018074",
"2018080",
"140059",
"140060",
"140062",
"140063",
"140068",
"140069",
"140072",
"140073",
"140075"
),
sp_rename = c(
rep("Northern elephant seal", 4),
rep("Southern elephant seal", 9)
),
lenmass = c(144, NA, 131, 142, 141, 141, 118, NA, 136, NA, NA, 134, NA)
),
by = c("sp_rename", "id_rename")
) %>%
# reorder column
relocate(nb_days, N, travel_distance, weanmass, lenmass, divetype_perc,
.after = id_rename
) %>%
# setup group row
gt(groupname_col = "sp_rename") %>%
# spanner (several columns into one column)
tab_spanner(
label = "Maximum depth (m)",
columns = c(
Maxdepth_mean,
Maxdepth_plus_minus,
Maxdepth_sd,
sparkline_qt_maxdepth
)
) %>%
tab_spanner(
label = "Dive duration (min)",
columns = c(
Dduration_mean,
Dduration_plus_minus,
Dduration_sd,
sparkline_qt_dduration
)
) %>%
tab_spanner(
label = md("Daily median drift rate"),
columns = c(sparkline_driftrate)
) %>%
tab_spanner(
label = md("Travel distance"),
columns = c(travel_distance)
) %>%
tab_spanner(
label = md("Mass"),
columns = c(weanmass)
) %>%
tab_spanner(
label = md("Length"),
columns = c(lenmass)
) %>%
tab_spanner(
label = md("Dive type proportions"),
columns = c(divetype_perc)
) %>%
# plot
gt_ggplot_sparkline(sparkline_qt_maxdepth, "sparkline_qt_maxdepth") %>%
gt_ggplot_sparkline(sparkline_qt_dduration, "sparkline_qt_dduration") %>%
gt_ggplot_driftrate(sparkline_driftrate, "sparkline_driftrate") %>%
gt_plt_bar_stack_extra(
divetype_perc,
width = 65,
labels = c("Transit", "Active Btt.", "Drift", "Benthic"),
palette = c("#000000", "#444444", "#888888", "#CCCCCC")
) %>%
# alignement
cols_align(
columns = c(N, Maxdepth_sd, Dduration_sd),
align = "left"
) %>%
cols_align(
columns = c(N, Maxdepth_mean, Dduration_mean),
align = "right"
) %>%
cols_align(
columns = c(travel_distance, weanmass, lenmass, Maxdepth_plus_minus, Dduration_plus_minus),
align = "center"
) %>%
# format
fmt_number(
columns = Maxdepth_mean,
decimal = 1
) %>%
fmt_number(
columns = ends_with("_sd"),
decimal = 1
) %>%
# rename columns
cols_label(
N = "# dives",
id_rename = md("ID"),
nb_days = "# days",
travel_distance = "(km)",
weanmass = "(kg)",
lenmass = "(cm)",
Maxdepth_mean = md("Mean"),
Maxdepth_plus_minus = md("±"),
Maxdepth_sd = md("SD"),
sparkline_qt_maxdepth = md("Trend"),
Dduration_mean = md("Mean"),
Dduration_plus_minus = md("±"),
Dduration_sd = md("SD"),
sparkline_driftrate = md("(m.s<sup>-1</sup>)"),
sparkline_qt_dduration = md("Trend")
) %>%
# add color square
text_transform(
locations = cells_row_groups(),
fn = function(x) {
# identify sp
sp <- unique(x)
# set colour
colour <-
if_else(grepl("Northern", sp), colours[1], colours[2])
# html to add the color box
purrr::map(x, ~ html(
glue(
"<div><span style='height: 15px;width: 15px;background-color: {colour};display: inline-block;border-radius:5px;float:left;top:13%;left:5%;'</span> <span style='display: inline-block;float:left;line-height:20px;padding: 0px 25px;white-space:nowrap;'>{sp}</span></div>"
)
))
}
) %>%
# color cols
gt_highlight_cols(
columns = c(
Maxdepth_mean,
Maxdepth_plus_minus,
Maxdepth_sd,
sparkline_qt_maxdepth,
sparkline_driftrate,
weanmass,
N
),
fill = "lightgrey",
alpha = 0.5
) %>%
# footnote
tab_footnote(
footnote = "Recorded",
locations = cells_column_labels(columns = c(N, nb_days))
) %>%
# color rows
opt_row_striping() %>%
# set horizontal padding for plus minus
tab_style(
style = "padding-left:0px;padding-right:0px;",
locations = cells_column_labels(columns = ends_with("plus_minus"))
) %>%
tab_style(
style = "padding-left:0px;padding-right:0px;",
locations = cells_body(columns = ends_with("plus_minus"))
) %>%
# set horizontal padding for plus minus
tab_style(
style = "padding-top:0px;padding-bottom:0px",
locations = cells_body(columns = starts_with("sparkline"))
) %>%
# settings
tab_options(
# # width table
table.width = pct(180),
# table.width = pct(150),
# padding = vertical space between rows
data_row.padding = px(3),
# horizontal scroll
container.overflow.x = T
)| ID | # days1 | # dives1 | Travel distance | Mass | Length | Dive type proportions | Maximum depth (m) | Dive duration (min) | Daily median drift rate | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (km) | (kg) | (cm) | Transit | Active Btt. | Drift | Benthic | Mean | ± | SD | Trend | Mean | ± | SD | Trend | (m.s-1) | |||
Northern elephant seal |
|||||||||||||||
| 2018070 | 229.5 | 22,390 | 5,948 | 132 | 144 | 305.5 | ± | 180.0 | 12.9 | ± | 3.0 | ||||
| 2018072 | 251.3 | 22,799 | 10,430 | 138 | NA | 331.6 | ± | 187.2 | 14.1 | ± | 3.1 | ||||
| 2018074 | 222.8 | 25,679 | NA | 119 | 131 | 231.7 | ± | 150.9 | 11.1 | ± | 2.5 | ||||
| 2018080 | 213.9 | 19,028 | 8,334 | 142 | 142 | 296.5 | ± | 146.7 | 14.5 | ± | 2.4 | ||||
Southern elephant seal |
|||||||||||||||
| 140059 | 185.3 | 1,823 | 7,356 | 118 | 141 | 179.5 | ± | 112.5 | 11.4 | ± | 1.9 | ||||
| 140060 | 192.4 | 1,866 | 8,897 | 112 | 141 | 188.9 | ± | 102.6 | 9.7 | ± | 1.7 | ||||
| 140062 | 152.3 | 1,574 | 5,864 | 85 | 118 | 135.7 | ± | 60.4 | 7.0 | ± | 1.0 | ||||
| 140063 | 157.5 | 1,570 | 7,804 | NA | NA | 175.1 | ± | 113.0 | 10.2 | ± | 1.9 | ||||
| 140068 | 196.9 | 1,904 | 8,328 | 125 | 136 | 165.8 | ± | 113.3 | 11.1 | ± | 1.9 | ||||
| 140069 | 216.3 | 2,123 | 8,865 | NA | NA | 174.0 | ± | 125.1 | 9.2 | ± | 2.1 | ||||
| 140072 | 154.9 | 1,555 | 8,857 | NA | NA | 148.6 | ± | 79.9 | 8.3 | ± | 1.3 | ||||
| 140073 | 174.3 | 1,692 | 7,094 | 102 | 134 | 190.5 | ± | 117.2 | 10.3 | ± | 2.0 | ||||
| 140075 | 142.3 | 1,451 | 6,608 | NA | NA | 132.5 | ± | 90.3 | 7.8 | ± | 1.5 | ||||
| 1 Recorded | |||||||||||||||
# # for export
# summary_table_es %>% gtsave_extra(
# "test_table_1.png",
# vwidth = 2000,
# vheight = 580,
# # cliprect = "viewport"
# )Title: Descriptive statistics and visual representations of the dataset’s first offshore foraging trip for each northern and southern elephant seal. For maximum depth (m; note the inverted y-axis) and dive duration (min), the trend represents the changes in the daily 95th percentile through time (solid gray line) associated with a linear regression (black dashes). For drift rate (m.s-1), the daily median was calculated to represent the evolution over time, with positive values in orange and negative in violet. The measurements of length and mass were made at weaning.
Extra
# by species
data_comp %>%
.[!is.na(lat), `:=`(
sunrise_today = maptools::sunriset(matrix(c(lon, lat), ncol = 2),
date,
direction = "sunrise",
POSIXct.out = TRUE
)$time,
sunset_today = maptools::sunriset(matrix(c(lon, lat), ncol = 2),
date,
direction = "sunset",
POSIXct.out = TRUE
)$time
), ] %>%
# calculation day-time length
.[, day_time := as.numeric(difftime(sunset_today,
sunrise_today,
units = "hours"
))] %>%
# calculation night-time length
.[, night_time := 24 - day_time] %>%
# calculate maxdepth and dduration
.[, .(
result_depth = paste(
round(mean(maxdepth), 1),
"±",
round(sd(maxdepth), 1)
),
result_duration = paste(
round(mean(dduration / 60), 1),
"±",
round(sd(dduration / 60), 1)
),
result_day_time = paste(
round(mean(day_time, na.rm = T), 1),
"±",
round(sd(day_time, na.rm = T), 1)
),
result_night_time = paste(
round(mean(night_time, na.rm = T), 1),
"±",
round(sd(night_time, na.rm = T), 1)
)
), by = .(sp_rename)] %>%
# add wean mass based on Weanling Dive Metada.xlsx
merge(., data.table(
id_rename = c(
"2018070",
"2018072",
"2018074",
"2018080",
"140059",
"140060",
"140062",
"140063",
"140068",
"140069",
"140072",
"140073",
"140075"
),
sp_rename = c(
rep("Northern elephant seal", 4),
rep("Southern elephant seal", 9)
),
weanmass = c(132, 138, 119, 142, 118, 112, 85, NA, 125, NA, NA, 102, NA)
) %>%
.[, .(result_mass = paste(
round(mean(weanmass, na.rm = T), 1),
"±",
round(sd(weanmass, na.rm = T), 1)
)),
by = sp_rename
],
by = "sp_rename"
) %>%
merge(., data.table(
id_rename = c(
"2018070",
"2018072",
"2018074",
"2018080",
"140059",
"140060",
"140062",
"140063",
"140068",
"140069",
"140072",
"140073",
"140075"
),
sp_rename = c(
rep("Northern elephant seal", 4),
rep("Southern elephant seal", 9)
),
lenmass = c(144, NA, 131, 142, 141, 141, 118, NA, 136, NA, NA, 134, NA)
) %>%
.[, .(result_len = paste(
round(mean(lenmass, na.rm = T), 1),
"±",
round(sd(lenmass, na.rm = T), 1)
)),
by = sp_rename
],
by = "sp_rename"
) %>%
merge(., data_comp[, .(travel_distance = sum(travel_distance, na.rm = T) / 1000),
by = .(.id, sp_rename)
] %>%
.[, travel_distance := na_if(travel_distance, 0)] %>%
.[, .(result_distance = paste(round(mean(
travel_distance,
na.rm = T
), 1), "±", round(sd(
travel_distance,
na.rm = T
), 1))), by = .(sp_rename)], by = "sp_rename") %>%
merge(., data_comp[, .(nb_days = as.numeric(difftime(max(date),
min(date),
units = "day"
))),
by = .(.id, sp_rename)
] %>%
.[, .(nb_days = paste(
round(mean(nb_days, na.rm = T), 1),
"±",
round(sd(nb_days, na.rm = T), 1)
)),
by = .(sp_rename)
], by = "sp_rename") %>%
gt() %>%
tab_spanner(
label = md("Dive"),
columns = c("result_depth", "result_duration")
) %>%
tab_spanner(
label = md("Length (days)"),
columns = c("result_day_time", "result_night_time")
) %>%
tab_spanner(
label = md("Weaning"),
columns = c("result_mass", "result_len")
) %>%
tab_spanner(
label = md("Travel"),
columns = c("result_distance")
) %>%
cols_label(
sp_rename = "Species",
result_depth = "Depth (m)",
result_duration = "Duration (days)",
result_day_time = "Day",
result_night_time = "Night",
result_mass = "Mass (kg)",
result_len = "Length (cm)",
result_distance = "Distance (km)"
) %>%
opt_row_striping() %>%
# settings
tab_options(
# # width table
table.width = pct(175),
# table.width = pct(150),
# padding = vertical space between rows
data_row.padding = px(3),
# horizontal scroll
container.overflow.x = T
)| Species | Dive | Length (days) | Weaning | Travel | nb_days | |||
|---|---|---|---|---|---|---|---|---|
| Depth (m) | Duration (days) | Day | Night | Mass (kg) | Length (cm) | Distance (km) | ||
| Northern elephant seal | 289.1 ± 171.8 | 13 ± 4.7 | 13.3 ± 2.4 | 10.7 ± 2.4 | 132.8 ± 10 | 139 ± 7 | 8237 ± 2242.6 | 229.4 ± 16 |
| Southern elephant seal | 167 ± 106.7 | 9.5 ± 4.7 | 13 ± 3.1 | 11 ± 3.1 | 108.4 ± 15.6 | 134 ± 9.5 | 7741.6 ± 1093.4 | 174.7 ± 24.7 |
